Multidomain protein folding
N-Terminal Domains in Two-Domain Proteins Are Biased to Be Shorter and Predicted to Fold Faster Than Their C-Terminal Counterparts
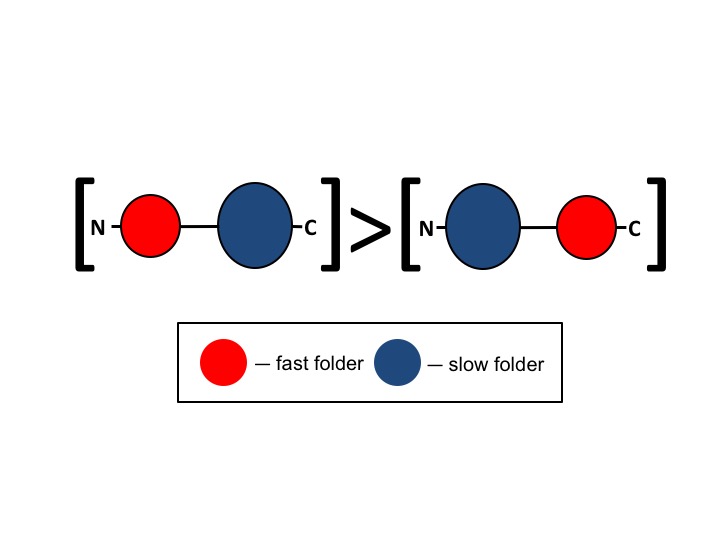
I revealed a strong tendency in all kingdoms of life for N-terminal domains in two-domain proteins to have shorter sequences than their neighboring C-terminal domains. Given that folding rates are affected by chain length, I asked whether the tendency for N-terminal domains to be shorter than their neighboring C-terminal domains reflects selection for faster folding N-terminal domains. Calculations of absolute contact order, another predictor of folding rate, provided additional evidence that N-terminal domains tend to fold faster than their C-terminal neighboring domains. A possible explanation for this bias is that faster folding of N-terminal domains reduces the risk of protein aggregation during folding by preventing formation of non-native interdomain interactions. This explanation is supported by protein expression analyses I performed which demonstrated that two-domain proteins with a shorter N-terminal domain are much more abundant than those with a shorter C-terminal domain. These findings, therefore, suggest a previously unrecognized mechanism for prevention of aggregation of neighboring domains in multi-domain proteins.
This work was published in Cell Reports.