Bio
I am a computational biologist affiliated to the Department of Data Sciences and the Department of Medical Oncology at Dana-Farber Cancer Institute, and the Broad Institute of MIT and Harvard. My main research focus is to understand complex domains that involve large amounts of uncertainty and noise using statistical models, machine learning and computer simulations.
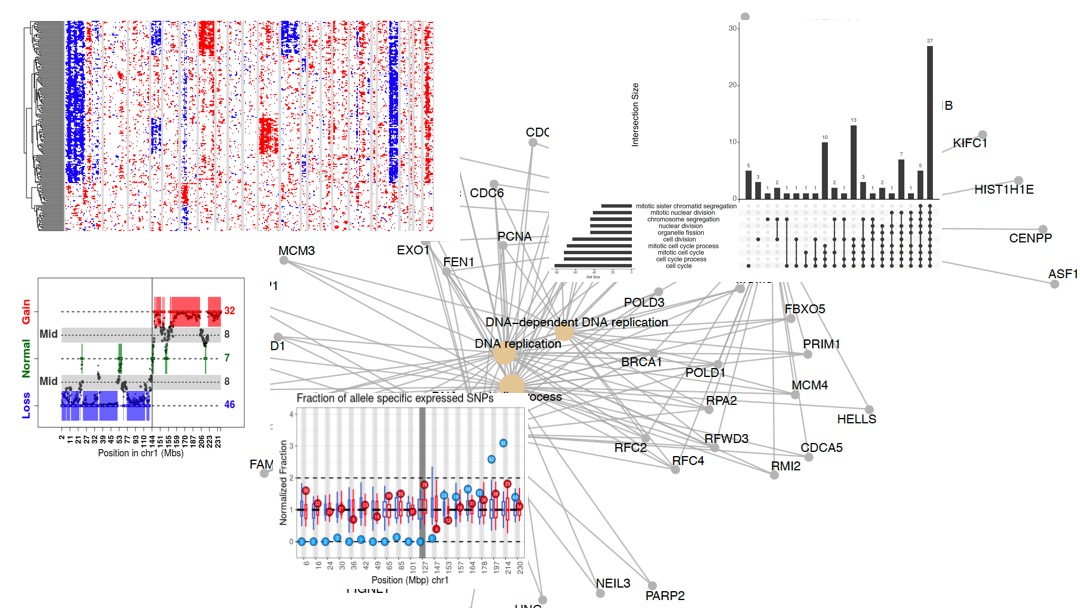
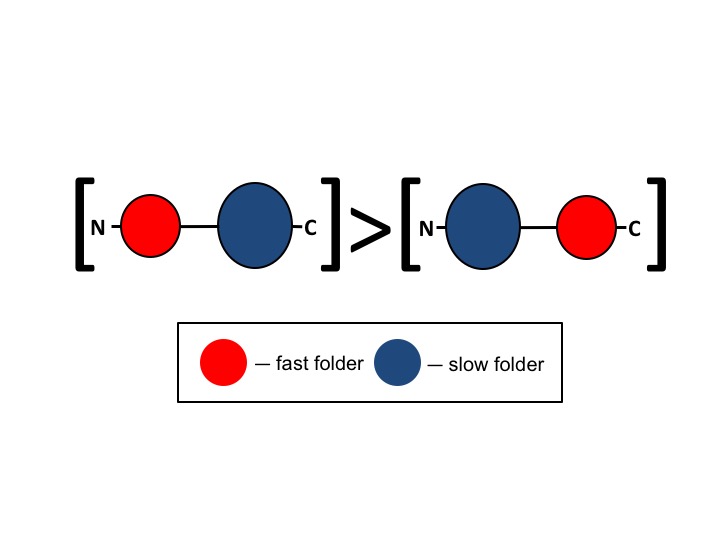
I like to be driven by real-world problems and therefore, most of my academic work tackles biological applications. Recently, I have focused on analyzing single cell RNA-seq data to study the effect of genome instability on RNA transcription and the impact of mutations on cancer and normal tissue developement. In my earlier work in the field, I focused more on theoretical aspects in biology. Using toy models and Monte Carlo simulations I demonstrated how certain features in protein structure could relate to the protein folding process. Later on, in my PhD work, I have focused on protein structure prediction using correlated mutation analysis, multi-domain protein folding and protein aggregation.
Before joining Dana-Farber Cancer Institute, I worked at Compugen, a clinical-stage drug discovery and development company (NASDAQ:CGEN). In compugen, I developed data mining infrastructre to integrate multi-omics data (e.g. RNA expression and ChIP-Seq data), supported new discoveries and performed on-going research of potential drug targets. I developed and utilized machine learning (e.g. Random Forests, Neural Networks and linear models) and pattern recognition methods to study protein sequence and structure (e.g. PDB, Uniprot), protein abundance and RNA expression (e.g. using microarray and NGS data along with Mass spectrometry data), biological function (e.g. using GO terms, MsigDB and PUBMED) and more.
I obtained my PhD in computational biology at the Weizmann Institute of Science and Bar-Ilan University (as a collaboration), Israel, in 2016.
Publications
Most recent publications on Google Scholar.
Adipocytes sensitize melanoma cells to environmental TGF-β cues by repressing the expression of miR-211.
Tamar Golan, Roma Parikh, Etai Jacob, Hananya Vaknine, Valentina Zemser-Werner, Dov Hershkovitz, Hagar Malcov, Stav Leibou, Hadar Reichman, Danna Sheinboim, Ruth Percik, Sarah Amar, Ronen Brenner, Shoshana Greenberger, Andrew Kung, Mehdi Khaled, Carmit Levy
Science Signaling, 23 Jul 2019: Vol. 12, Issue 591, eaav6847
Breaching self-tolerance to Alu duplex RNA underlies MDA5-mediated inflammation.
Sadeem Ahmad, Xin Mu, Fei Yang, Emily Greenwald, Ji Woo Park, Etai Jacob, Cheng-Zhong Zhang, Sun Hur
Cell, Volume 172, Issue 4, 8 February 2018, Pages 797-810.e13
Cross-linking reveals laminin coiled-coil architecture.
Gad Armony, Etai Jacob, Toot Moran, Yishai Levin, Tevie Mehlman, Yaakov Levy, Deborah Fass
PNAS, November 22, 2016 113 (47) 13384-13389
Extracting Insights Into Protein Structures from Their Sequences.
Etai Jacob
Dissertation, Feb. 2016
Codon-level information improves predictions of inter-residue contacts in proteins by correlated mutation analysis.
Etai Jacob, Ron Unger, Amnon Horovitz
eLife, Sep 2015, 4:e08932.
N-terminal domains in two-domain proteins are biased to be shorter and predicted to fold faster than their C-terminal counterparts.
Etai Jacob, Ron Unger, Amnon Horovitz
Cell Reports, Volume 3, Issue 4, 25 April 2013, Pages 1051-1056
Computational studies of protein chaperone mediated folding interactions.
Etai Jacob
M.Sc. Thesis, 2007
Different mechanistic requirements for prokaryotic and eukaryotic chaperonins: a lattice study.
Etai Jacob, Amnon Horovitz, Ron Unger
Bioinformatics, Volume 23, Issue 13, July 2007, Pages i240–i248
A tale of two tails: why are terminal residues of proteins exposed?.
Etai Jacob, Ron Unger
Bioinformatics, Volume 23, Issue 2, 15 January 2007, Pages e225–e230
New cell-penetrating peptides and uses thereof.
Etai Jacob, Amir Toporik, Ronen Shemesh, Ofer Levy
Application US13/976,502 filed by Compugen Ltd., 2010-12-27
Research
Resume
-
Dana-Farber Cancer Institute Sep 2016-nowComputational Biologist
Department of Data Sciences -
Compugen 2011-2016Senior Scientist
Computational Research and Discovery -
Weizmann Institute of Science
Bar-Ilan University 2011-2016Ph.D. Candidate
Computational Biology -
Compugen 2008 - 2011Project leader
Computational Research and Discovery -
Compugen 2006 - 2008Algorithm developer
Algorithmic Research group -
Bar-Ilan University 2005 - 2006Teaching assistant
Introduction to computing and computer aided problem solving -
Bar-Ilan University 2005 - 2006M.Sc. student
Computational biology